Also the species subdivision still needs to be clarified. In this review, by Ibrahim Diler et al, Isparta, Turkey, studies related with gilthead sea bream populations and problems with the clarification of gilthead seabream population structure in hatchery and wild were reviewed. Also, it was intensified on potential research areas, having importance in determination of population structure of gilthead seabream such as morphology and behaviour in hatchery and wild. Determination of the genetic structure of S. aurata populations based on behaviour and morphology will not only contribute greatly to biological clarification of the species but also will help the development strategy for conservation of natural stocks (e.g., protecting the stopover points) and the determination of reference stocks, which is important for the culture of gilthead seabream to use in the genetic improvement programmes.
Introduction
The gilthead seabream (Sparus aurata Linnaeus 1758) is a perciform fish, belonging to the family Sparidae and to the genus Sparus. It inhabits the Atlantic coasts of Europe, Mediterranean and Black sea (rare) and is one of the most important marine fish in fishery and aquaculture (especially in the Mediterranean area). Due to its euryhaline and eurythermal habits, the species is found in both marine and brackishwater environments such as coastal lagoons and estuarine areas, in particular during the initial stages of its life cycle. The gilthead seabream is usually found on rocky and seaweed bottoms but it is also frequently caught on sandy grounds. Young fish remain at low depth (>30 m), whereas adults can reach deeper waters (maximum depth of 150 m) (Moretti et al., 1999). It is mainly carnivorous (shellfish including mussels and oysters), accessorily herbivorous (Froese and Pauly, 2006). It is a highly fecund protandrous hermaphrodite. Ovarian development is asynchronous and exhibits mass spawning (daily and for a period of >3 months) behaviour. Breeding season is usually ranging from October-December.
The gilthead seabream is a functional male in the first 2 years and at sizes over 30 cm become females. After spawning, the eggs, which are spherical and transparent have a diameter of slightly <1 mm and present a single large oil droplet. The gilthead seabream is regularly present on the markets in Adriatic, Greece, Turkey and Maghreb. It is commercialised fresh, refrigerated and frozen. Trawl nets, bottom set longlines, hand lines are also commonly used in fishing of gilthead seabream (Moretti et al., 1999). Before seabream aquaculture started, the species was reared traditionally in lagoons all along the Mediterranean coast. As with seabass, seabream domestication is recent and started at the same time with the establishment of reproduction and husbandry methods in the 1980s. The first breeding programmes took place in France and Israel in the 90s. A high proportion of seabream fingerlings produced today is derived from unselected broodstocks (Chavanne et al., 2008). Sea breams are farmed extensively in lagoons or intensively in tanks, cages or earthen ponds (Turkey). At present, most production is from intensive farming, with average densities of 20-100 kg m -3 and a FCR is 1.5-2:1. Extensive farming still remains a traditional activity in some regions but with a very low impact on the market. At the beginning of the 1990s, twenty sea bream hatcheries were operating in the Mediterranean; at 2006, over 65 hatcheries are distributed in Croatia, Cyprus, France, Greece, Italy, Morocco, Portugal, Spain and Tunisia (Sola et al., 2006). Seabream production reached >128,943 tonnes in 2008 with major contributions from Greece (47 per cent), Turkey (21 per cent), Spain (19 per cent) and Italy (7.4 per cent) (FEAP, 2008).
Materials and Methods
Population structure of the gilthead seabream: The commercial importance of gilthead seabream has generated intense interest in its molecular genetics. Studies on gilthead sea bream and its populations have been c arried out through gene-enzyme systems (Cervelli et al., 1985; Palma et al., 2001; Alarcon et al., 2004; Ben-Slimen et al., 2004; Rossi et al., 2006), AFLP (Miggiano et al., 2005), mtDNA (Funkenstein et al., 1990; Magoulas et al., 1995; Alarcon et al., 2004), RAPD (Bilgen et al., 2007) and microsatellite analyses (Batargias et al., 1999; Palma et al., 2001; Launey et al., 2003; Alarcon et al., 2004; Miggiano et al., 2005; Brown et al., 2005a; De Innocentiis et al., 2004, 2005). Cervelli et al. (1985) studied on natural populations gilthead seabream from Southern Sicily and Northern Adriatic Sea using allozyme markers. Funkenstein et al. (1990) first reported on mitochondrial DNA polymorphism of gilthead seabream broodstocks in Israel by using RFLP analysis of the whole mitochondrial DNA (mtDNA) molecule. Magoulas et al. (1995) reported on mtDNA polymorphism for Greece broodstocks. Alarcon et al. (2004) compared cultivated and wild seabream stocks from the Atlantic and Mediterranean coasts using allozymes, microsatellite and mtDNA markers. De Innocentiis et al. (2004) investigated the genetic variability of gilthead seabream populations from the Atlantic Ocean, Mediterranean Sea and Adriatic Sea using microsatellite marker. Alarcon et al. (2004) analysed six wild and five cultivated sample sets covering the South Atlantic and Mediterranean European using Microsatellite, allozyme markers and mtDNA variation. Ben-Slimen et al. (2004) reported strong genetic differentiation between populations of gilthead seabream along the Tunisian coast. Rossi et al. (2006) studied on the genetic structure of gilthead seabream in the Central Mediterranean Sea using allozyme markers. Bilgen et al. (2007) studied on the genetic diversity of gilthead Seabream broodstocks from wild in Turkey by using RAPD markers and suggested there was some genetic exchange between the Aegean Sea and eastern Mediterranean wild seabream populations.
The first studies reported conficting data concerning the existence of panmictic (Cervelli et al., 1985) or subdivided populations (Funkenstein et al., 1990). More recent studies (Alarcon et al., 2004; Ben-Slimen et al., 2004; De Innocentiis et al., 2004; Rossi et al., 2006; Bilgen et al., 2007) have depicted a picture of species subdivision but this still needs investigation.
Ben-Slimen et al. (2004) reported that strong differentiation (Total Fst = 0.093, highest pairwise Fst = 0.265; for marine fish, mean Fst = 0.062 has been detected between samples from six localities from the northern and southern Tunisian coasts using allozyme markers. Alarcon et al. (2004) reported that the wild populations from Atlantic and Mediterranean revealed a slight degree of differentiation (Fst for allozymes; 0.031 and for microsatellites; 0.036) but not apparently associated with geographic or oceanographic factors (Alarcon et al., 2004). Rossi et al. (2006) reported that a hierarchical analysis of population subdivision made it possible to identify three different assemblages found (Fst = 0.017) in the Adriatic Sea, Tyrrhenian Sea and Sardinian Channel, though an isolation by distance model could be rejected. De Innocentiis et al. (2004) found a slight but significant population structure (Fst = 0.01) for the Western Mediterranean Sea (Sardinian Sea, Sardinian Channel and Central Tyrrhenian Sea), Atlantic Ocean and the Adriatic Sea. When evaluated the results of above studies together, species subdivision still needs to be clarified. There have been some granted EU projects concerning the gilthead seabream so far such as Bridgemap, Marine Genomics Europe (MGE), Aquafirst, Fine Fish, Geneimpact and Aqua Breeding. It was focused on gilthead seabream genome with MGE, Bridgemap and Aquafirst. Through, the MGE, many EST (Expressed Sequence Tags), SNP (Single Nucleotide Polymorphism), SSR-EST (sequences containing tandem repeats) were identified. In Bridgemap, some of the results were published (Senger et al., 2006). They reported the production of a seabream whole-genome Radiation Hybrid (RH) panel in which quality was ascertained by the construction of a 2-Mb-resolution RH map. The map encompassed 440 markers (288 microsatellites, 82 gene based markers and 70 STS) suitable for linkage analysis and comparative mapping studies. The genetic origin of malformations in gilthead sea bream fry was studied with Fine Fish project. Within the Geneimpact project, the genetic impact of aquaculture activities on native populations is being evaluated.
Interaction between wild and cultured stocks: Restocking programs, accidental escapes of fish from farms, spawning of females in cages represent potential threat to the genetic integrity of natural populations. But not much is known about their impact on wild populations.
Alarcon et al. (2004) compared the wild and cultured populations and reported that cultivated populations show a slight decrease of variability related to the wild ones but was not sufficient to document inbreeding depression effects and that the cultured seabream were genetically very different from the local wild ones and there was no evidence for significant genetic flow between them.
Genetic improvement of the gilthead seabream: There has been an increasing interest in the genetic improvement of gilthead seabream as there has been only a short history of captive management and evidence of large potential gains for selective breeding of certain traits (Batargias, 1998; Knibb et al., 1997). Genetic response in gilthead seabream ranges between 5 and 10 per cent per generation according to industrial reports (Knibb, 2000). From some more recent programmes in Greece, the estimated selection response for growth rate resulting from the first cycle of selection was 20, 22 per cent (Sola et al., 2006; Chavanne et al., 2008).
Results and Discussion
Problems related with determination of the structure of gilthead seabream populations in wild: Some problems with clarification of the structure of natural populations of gilthead seabream are as follows: it should be urgently need to be investigated the life cycle and ecology of the species in the wild because there is little information on its biology, in particular the effective and census sizes, spatial distribution, spawning grounds and behaviour and eventual migration (Sola et al., 2006). The structure of wild seabream populations is still unclear. Subdivision of the species needs to be clarified, in particular through further analysis covering the whole species distribution and also seasonal or year to year variation (Chavanne et al., 2008). There is a lack of knowledge on the effects of marine currents or on the migratory behaviour of S. aurata adults ( Rossi et al., 2006; De Innocentiis et al., 2005). Small scale studies, in spite of their limited geographic extent, might be useful in detecting local peculiar situations (Rossi et al., 2006). Alarcon et al. (2004) reported that their study provided an insight into the population structure of S. aurata, although more questions have arisen that need to be solved and this could be achieved by further screening of small-scaled targeted sample sets in the studied area. Data available in the literature lead us to conclude that the South Atlantic and Mediterranean populations of S. aurata present some sort of structuring pattern, although it is lower than the average of marine species which cannot be associated with geographic and/or oceanographic known factors. This undefined structuring sharply contrasts with the high geographic differentiation detected in other sparid species which are very close to S. aurata, in terms of biological and ecological traits. The actual reasons of such structuration are unknown, however they might be associated with fluctuations in the effective population size and/or bottlenecks and expansions, possibly combined with differences in gene flow rates (Alarcon et al., 2004).
Hatchery juveniles which originated from broodstocks of often unknown origin are released into coastal lagoons to enhance natural stocks. Thus, accidental escapes from floating cages or restocking programs may represent a potential threat for the genetic diversity of local populations, i.e., the genetic integrity of the species. A proper management of the species, therefore, depends on the knowledge of its population structure ( Rossi et al., 2006; De Innocentiis et al., 2004). Gilthead seabream juveniles are frequently exploited illegally from wild sources for decreasing the juvenile cost in culture. Spawning a large quantity of eggs by gilthead seabream females in the cages may affect genetic structure of wild stocks (Chavanne et al., 2008). A proper management plan based on breams biology and population structure for conservation of the wild stocks is needed.
Problems related with gilthead seabream broodstock populations in hatchery: Some problems with the broodstock populations of the gilthead seabream in hatchery are as follows: a major problem in the aquaculture of sea bream is the variation in genetic for the farmed stocks regarding the influence of the culture practice and/or the biology of the species itself. However, seabream farmers have not released records for the stocks under study on both the constitution and maintenance of broodstocks. For example, some farms buy their fry from wholesale suppliers whereas others produce their own fry using breeding stock which is not maintained under any guidelines. In general, the cultivated populations are supported with wild fish taken from a neighbouring area. The history, the origin, the size and the sex ratio composition of the breeding stocks are generally not suitably controlled (Alarcon et al., 2004). In species where sexes are separated and unchanged through life, farmers regularly add new individuals of the most vulnerable sex. Because of the protandric hermaphroditism shown by S. aurata, the farmers may depend on the same individuals to act initially as males and later as females. As a consequence, everything else being equal, the effective population size of seabream breeding stocks would be smaller than that of non-hermaphrodite species. This aspect, coupled with the putative small numbers of founders derived from wild, could result in inbreeding depression (Alarcon et al., 2004).
Gilthead seabream is a highly fecund hermaphroditic species that exhibits mass-spawning behaviour. This gives the opportunity for only a few individuals to be used as broodstock in the production of a cohort of juveniles. With this type of mating system, large variation in individual broodstock contribution to offspring is expected. This will cause lower effective population sizes than the actual census. Under this situation the risk of inbreeding becomes especially high in cultured populations of this species (Brown et al., 2005b). For the fish with high fecundity such as seabream, important nongenetic maternal effects and high mortality at early life stages can rapidly induce a loss of genetic variability in selected populations if there is no control of mating schemes (Perez-Enriquez et al. 1999).
To establish a breeding programme, the long-term sustainability of the mating system and the management of inbreeding in the population under selection need to be considered. In commercial production systems of gilthead seabream, there is often no control over the mating system rather than to regulate the size of the broodstock groups. Batches of offspring from individual broodstock groups (50-60 fish) may use to form the bulk of selected populations because they can be maintained as units via the farming system, with the benefit of minimising environmental variance and competition effects during development (Brown et al., 2005b). There is no clear information on the contribution of individual fish to the offspring is usually unknown in gilthead seabream commercial farms and the only mating variables under control are the size and sex composition of the broodstock (Castro et al., 2007). Genetic aspects of fish stock management are not very well established methodologies in Mediterranean aquaculture yet. Many farmers are raising stocks that are either wild or semi wild (only a few generations removed from the wild). Managers concerns arise mainly from environmental aspects of management (nutrition, water quality management, disease control and prevention, etc.) because so rapid improvement may be obtained, ignoring the genetic aspects in the early stages of culture. Indeed genetics requires sophistication, well-planning, extra facilities and adds additional financial burdens. Unfortunately, many fish stocking programs have not been successful and mainly due to improper broodstock management in the hatcheries in the short or long term (Borrell et al., 2007).
Genetic differentiation might increase over time because of small effective population sizes in many farms and the restricted gene flow among them (Alarcon et al., 2004). There is need to defined reference seabream stocks for setting up suitable guidelines for founding and maintaining of cultivated stocks. Strain comparison studies, performance tests lack for gilthead seabream.
Potential research areas related with determination of population structure of Sparus aurata: In this section, based on the preliminary studies and observations, it was intensified on potential research areas, having importance in determination of population structure of gilthead seabream such as morphology and behaviour in hatchery and wild.
Behaviour
In wild: The gilthead seabream form schools with different characteristics. These schools stop over certain places at the coast at different seasons of the year and/or pass through certain routes. The number, average size and the phenotypes of individuals in the schools are different. Some characteristics of the schools formed by gilthead seabream on the Turkish coastal region of Aegean Sea are summarized on the Table 1. As shown in Table 1, the different characteristics of schools excite us to handle the information on the gilthead seabream populations in a different manner. However, it is not clear that whether this fish schools belong to the different populations or not. Level of gene flow among them is also not known.
In hatchery: Social and behavioural harmony was generally seen in breeding stocks of gilthead seabream.
Table 1: Some characteristics of the schools formed by gilthead seabream on the Turkish coastal region of Aegean Sea
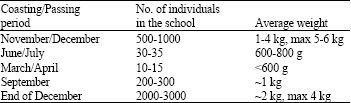
Towards to spawning period (approximately 2-3 weeks), one or two leader candidates may occur in a new breeding stock in hatchery and harmony impairs transiently till to clearing of the leader. The leader candidates of stock are males and have dark vertical bands. Size is not important for leadership. A small male also may be a leader candidate. If two leader candidates occur in a breeding stock, fight begins till one wins. Each individual in the stock obey the winning leader. The loosing leader candidate was rejected by school. Feeding of the lost leader candidate was blocked and was beaten by individuals in the school. It dies from starvation and/or injuries. We have isolated the lost leader candidate in a separate tank and fed him to gain condition during a month.
When we put back the conditioned leader candidate to tank, it was rejected again by school. Founding of a new breeding stock, adding of new individuals to breeding stock and/or mixing of the breeding stocks from different sources generally result in appearance of new leader candidates. After establishment of the leadership in a breeding stock, leadership problem wasn't seen, unless renewing of broodstocks and/or mixing of different breeding stocks was occurred. We monitored a breeding stock after clearing of the leader throughout the six years but we didn't observe any fight for leadership. For determining the mechanism of leadership and effects on breeding stock in hatchery and/or wild stock, there is a need to further studies.
Morphology: In the previous preliminary studies, we have observed a morphological difference between the groups of gilthead seabream juveniles collected from two different regions. The morphological difference was that mandibula was shorter compared with maxilla in some individuals while was almost identical in some individuals. The difference is seen on the Fig. 1. It seemed that social harmony was better and cannibalism was lower in the group with a greater number of fish having shorter mandibula compared with other group.
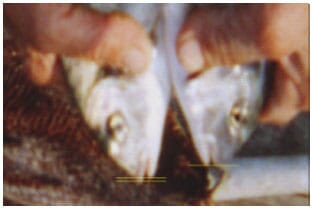
However, we didn't get the opportunity to restudy to clearly determine the relation between growth rate and morphological difference. Further studies are needed to clarify the relations among morphology, growth performance and population structure in gilthead seabream.
Conclusion
In the studies, to determine the population structure of the gilthead seabream so far, the samples have been collected from different and remote countries (from Atlantic to Eastern Mediterranean) as much as possible. In some other studies, wild and culture samples were compared and therefore a reliable data on the condition of the population was tried to achieve. Some early studies are related with determining of which broodstock used in the hatchery belong to which natural population. However, the information on the behaviour, morphology and growth performance related with genetic structure is lacking. Thus, determining genetic structure of S. aurata populations based on behaviour and morphology is necessary. This will not only contribute greatly to biological clarification of the species but also will help the development strategy for conservation of natural stocks (e.g., protecting the stopover points) and the determination of reference stocks, which is important for the culture of gilthead seabream to use in the genetic improvement programmes.
November 2013


