The White spot syndrome virus (WSSV) and the Penaeus stylirostris penstyldensovirus 1 (PstDV1) are major viral pathogens of penaeid shrimp, and pose a significant threat to shrimp aquaculture worldwide. Highly infectious, the WSSV has a large broad of hosts (more than 93 arthropods species described), and a cosmopolitan distribution, being reported in the majority of shrimp farming countries. The WSSV outbreaks in the whiteleg shrimp Litopenaeus vannamei were characterized by high mortality rates, reaching until 100% mortality in 3 to 10 days. By contrast, the PstDV1 causes a chronic disease in L. vannamei called runt deformity syndrome (RDS). The RDS causes cuticular deformities and retards growth, leading to lower production efficiency and reducing the market value of harvests by 10% to 50%.
Several methods, including histological examination, electron microscopy, in situ hybridization, and polymerase chain reaction (PCR) methods, have been developed for diagnosis of the WSSV, and the PstDV1 infections. Real-time PCR (qPCR) assays have been developed to detect the WSSV and the PstDV1. These qPCR methods have demonstrated higher sensitivities than those of the other diagnostic methods used for the detection of these shrimp viruses.
Currently, 53 scientific manuscripts and 11 patents describe different methods to diagnose WSSV according to Web of Science™ Database (Thompson Reuters, USA). However, there is no available qPCR assay for simultaneous detection of the WSSV and the PstDV1 in shrimp. A duplex qPCR method allows the simultaneous detection of two viruses in the same sample, which is more cost-effective than assaying for each virus separately. It is particularly interesting in epidemiological surveys, surveillance, and eradication programs, which require the analysis of large number of samples. Methods less time consuming and inexpensive are essential for the economic and technical feasibility of these programs, mainly when they are supported by governments. In addition, a duplex qPCR assay could improve the diagnosis of co-infection cases caused by WSSV and PstDV1. It is a growing issue to the biosecurity of shrimp farming.
The aim of this study was to develop and standardize a duplex qPCR assay for the simultaneous detection of the WSSV and the PstDV1 in clinical samples of diseased L. vannamei. In addition, to evaluate the performance of the TaqMan Universal PCR Master Mix (TUMM; Applied Biosystems, Carlsbad, CA, USA) and the QuantiTect Virus (QVK; Qiagen, Chatsworth, CA, USA) master mix with regard to the clinical sensitivity of the qPCR assay, as well as, different methods for qPCR results evaluation.
Results
qPCR Standardization
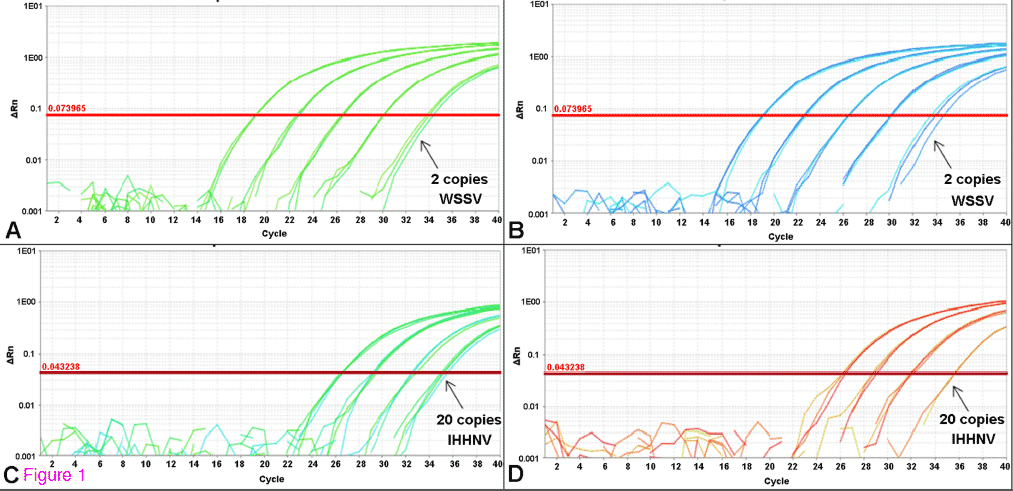
We standardized the duplex qPCR assay for detecting WSSV and PstDV1 in clinical samples of diseased whiteleg shrimp. The optimal primer-probe concentrations were 60 pmol of each primer and 25 pmol of probe per reaction for both the singleplex and duplex qPCR assays. No difference in the amplification of the standard curve was observed between the duplex and singleplex assays (Figure 1), regardless of the master mix used. The mean (n = 3) PCR efficiencies of the singleplex (WSSV = 91%, PstDV1 = 90%) and duplex (WSSV = 91%; PstDV1 = 101%) assays that used the TUMM were within the optimal recommended range. The mean (n = 3) PCR efficiencies for the singleplex (WSSV = 96%, PstDV1 = 98%) and duplex (WSSV = 109%, PstDV1 = 109%) assays that used the QVK master mix were also within the optimal recommended range. The detection limits were two copies of WSSV control plasmid and 20 copies of PstDV1 control plasmid per reaction for both the singleplex and duplex assays (Figure 1).
Figure 1 Amplification plots of standard curves of singleplex and duplex qPCR.
Standard curves of serial dilution from 2 × 104 to 2 copies of control plasmids showing the detection limit of two copies for WSSV in singleplex (A) and duplex format (B), and 20 copies for PstDV1 in singleplex (C) and duplex format (D).
The potential cross-interaction of the different DNA templates and its interference with PCR efficiency were evaluated using a 6 × 6 checkerboard analysis. The amplification plots of the WSSV and PstDV1 control plasmids were not significantly affected in mixtures with concentrations between 2 × 101 and 2 × 104 copies per reaction. At the highest PstDV1 plasmid concentration (2 x 105 copies per reaction) the detection limit of the WSSV plasmid increased from two to 20 copies. By contrast, the detection limit of the PstDV1 plasmid was not affected by any of the WSSV concentrations tested.
Clinical sensitivity
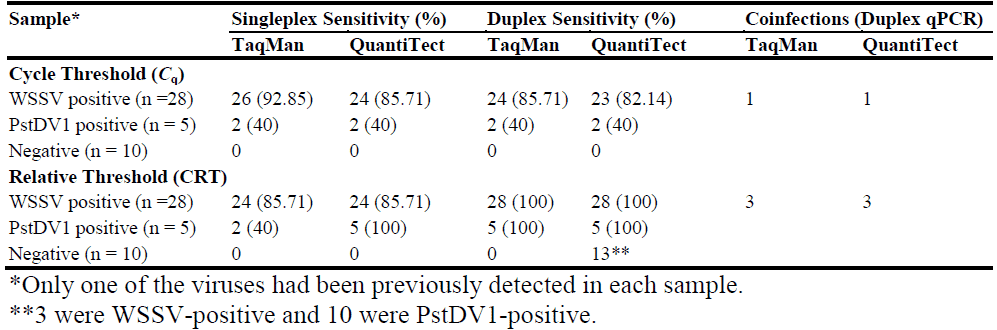
In the cycle threshold evaluation, no significant difference (P = 0.9265) in clinical sensitivity or specificity was observed between the singleplex assays for WSSV and PstDV1 and the duplex assays that used the same master mix (Table 1). The TUMM assays demonstrated superior clinical sensitivity for WSSV detection. However, no significant difference in sensitivity for PstDV1 detection was observed between the TUMM and QVK master mixes, with both products demonstrating a feasible clinical sensitivity for PstDV1 detection. The standardized duplex qPCR confirmed the presence of viral DNA in 28 samples that had previously tested positive for WSSV or PstDV1. The Cq values in the TUMM assay were lower than those for the QVK assay. An average reduction in Cq values of 3.11 was confirmed in the WSSV singleplex assay, and a 4.98 average reduction in Cq values was observed in the IHHNV singleplex assay. The TUMM duplex qPCR assays for WSSV and PstDV1 had 2.89 and 4.83 lower average Cq values, respectively. One case of coinfection was identified.
Table 1 Clinical sensitivities of qPCR assays for the detection of the WSSV and the
PstDV1
The use of the relative threshold algorithm for data analysis significantly increased the clinical sensitivity of PstDV1 detection (P = 0.0026; Table 1). However, no significant difference in sensitivity for WSSV detection was observed (P = 1.0). Using the relative threshold algorithm for data analysis, the singleplex and duplex QVK assays demonstrated a higher level of clinical sensitivity (100% for both viruses) than the TUMM assays, compared with those determined using the cycle threshold algorithm. However, 13 false-positive results, 3 false-positive for WSSV and 10 false-positive for PstDV1, were obtained in the QVK duplex qPCR assay. No significant difference in clinical sensitivity was observed between the singleplex and duplex QVK qPCR assays. However, the use of the relative threshold algorithm for the analysis of the data for the QVK duplex qPCR reduced the specificity of the assay from 100% to 43.47%, compared with the specificities obtained using the cycle threshold algorithm. Three coinfections were identified in both the QVK and TUMM duplex assays using the relative threshold algorithm.
Discussion
A duplex qPCR was standardized for WSSV and PstDV1 detection in clinical samples using primers and probes that have been previously validated. Specificities and sensitivities similar to those of the singleplex assays were obtained using the optimized duplex qPCR conditions, allowing the identification of both singular and coinfections in diseased whiteleg shrimp. The simultaneous identification of the WSSV and the PstDV1 is useful because such coinfections in shrimp are common.
The analytical sensitivity of the duplex qPCR assay was similar to those singleplex for WSSV and PstDV1 assays and those obtained in previous studies. The IHHNV detection limit of standardized duplex qPCR assay was similar to that reported for a real-time multiplex PCR method in a previous study. However, this duplex qPCR assay detected 10- and 627-fold lower amounts of PstDV1 DNA than the previously described conventional duplex and multiplex PCR methods. In addition, the WSSV detection limit for duplex qPCR assay was 1-fold lower than that obtained using previously described multiplex PCR methods, and 10 copies lower than that obtained using a previously described multiplex qPCR method. These data demonstrate the superior performance of standardized duplex qPCR method, compared with other conventional and real-time PCRbased methods. In addition, the lower detection limit of standardized duplex qPCR assay makes it a clinically useful tool for identifying animals with a low viral load, such as that observed in shrimp larvae and post-larvae , reducing the potential for false negative results.
Data from qPCR analyses have commonly been evaluated using a fit point method, which uses a fixed threshold based on the difference in baseline fluorescence intensities to determine the Cq value of each sample. The qPCR efficiency is affected by multiple factors that can lead to significant quantification problems. Such confounders are particularly relevant to qPCR-based clinical diagnostics for aquatic animal diseases because some inhibitors can contaminate the DNA samples from the tissues of certain species. Sigmoidal-fit models can reduce these types of inaccuracies because their algorithms analyze each strain separately.
In the present analysis of the various qPCR methods, the use of the relative threshold algorithm, which is based on a sigmoidal-fit model, substantially improved the performance of both the singleplex and duplex qPCR assays, and the clinical sensitivity was increased from 40% to 100%. Inhibitors of qPCR are a common issue in diagnostics for crustaceans, leading to frequent misdiagnoses. The superior performance of TUMM duplex qPCR assay should resolve such problems with regard to the detection of the WSSV and the PstDV1, while reducing false negative results. The QVK master mix contains an optimized combination of potassium chloride, ammonium sulfate, and a synthetic factor that enhance primer annealment. However, this may also cause nonspecific annealing in some cases, the type of which the present results and those of previous studies have confirmed.
Using the duplex qPCR assay, three cases of WSSV- PstDV1 coinfection were identified, for which the animals presented with clinical signs of one disease only. Using a duplex PCR method, Yang et al. were the first to describe a WSSV- PstDV1 coinfection in L. vannamei. Xie et al. also identified one case of WSSV- PstDV1 coinfection among 15 animals that were screened using a multiplex qPCR method. The standardized duplex qPCR assay identified a higher frequency of WSSV- PstDV1 coinfections than those reported in previous studies, which might be attributed to its superior sensitivity, compared with that of these previously described methods. This may also indicate that the frequency of natural coinfection in Brazilian shrimp farms is higher than that of other countries. Previous studies demonstrated that WSSV and PstDV1 infections occurred in shrimp cultivated in Brazil and other countries of South America. However, future studies have to be performed to address the frequency of WSSV/ PstDV1 coinfection in shrimp farms of the region.
Conclusions
The standardized duplex qPCR is a robust and highly sensitive diagnostic tool for the simultaneous detection of the WSSV and the PstDV1 in whiteleg shrimp. The use of the TUMM and the relative threshold method of data analysis in WSSV/ PstDV1 duplex qPCR method provided optimal levels of sensitivity and specificity.
Competing interests
The authors declare that they have no competing interests.
July 2014