Rapid and precise detection of viral hemorrhagic septicemia virus (VHSV) is important for disease prevention and containment by reducing the risk of spread or movement from an infected zone or compartment.
Currently, the internationally accepted method for diagnostic, surveillance, and confirmation testing is virus isolation from fish tissue specimens performed on bluegill fry (BF-2), epithelioma papulosum cyprini (EPC), and fathead minnow (FHM) cell cultures with follow-up antibody or nucleic acid based virus identification (OIE 2009).
Multiple real-time reverse transcription polymerase chain reaction (rRT-PCR) assays have been developed in order to reduce the amount of time required to obtain a test result. The use of rRTPCR as a robust targeted surveillance and diagnostic tool is becoming more widely accepted in both aquatic and terrestrial animal health management.
In the USA, VHSV genotype IVa is documented in fish collected from waters in the Pacific Northwest (Hershberger et al. 2010), while VHSV genotype IVb is documented in fish collected from the waters of the Laurentian Great Lakes (Faisal et al. 2012).
Many other regions or watersheds in the USA are free from VHSV. The emergence of VHSV IVb in the Laurentian Great Lakes of North America triggered intense surveillance efforts and disease investigations, and VHSV is now considered enzootic in these wild fish populations.
Similarly, VHSV is often considered enzootic in the Pacific Northwest, even though the actual disease incidences are unknown.
VHSV represents a serious risk to farmed or managed fish populations within the Great Lakes watershed and the Pacific Northwest. In addition, dissemination risks are a consideration when moving fish from these areas to other regions within or outside of the USA (Gustafson et al. 2010, VHSV Expert Panel and Working Group 2010).
A rapid and high-throughput diagnostic assay would be extremely useful for controlling VHSV and for facilitating fish trade. However, as testing for VHSV occurs in state, federal, and private laboratories located across the USA and this information is used for disease management, risk analysis, and regulatory purposes, it is imperative to adopt a standardized testing protocol.
In this manuscript, we report on a collaborative side-by-side evaluation of the TaqMan® based assay developed by Jonstrup et al. (2013; hereafter ‘Jonstrup assay’) and the 1-step format of Phelps et al. (2012; ‘Phelps assay’) that utilizes primers and probe developed by Garver et al. (2011) in an effort to estimate the diagnostic sensitivity and specificity of each test procedure.
The 2 assays were selected based on a comparison of analytical performance of 4 assays (Warg et al. 2014, this volume). Eight laboratories evaluated a tissue panel that contained 200 VHSV virus isolation positive and 200 VHSV virus isolation negative tissue homo genate supernatants using standardized rRT-PCR testing protocols.
Results
Diagnostic sensitivity
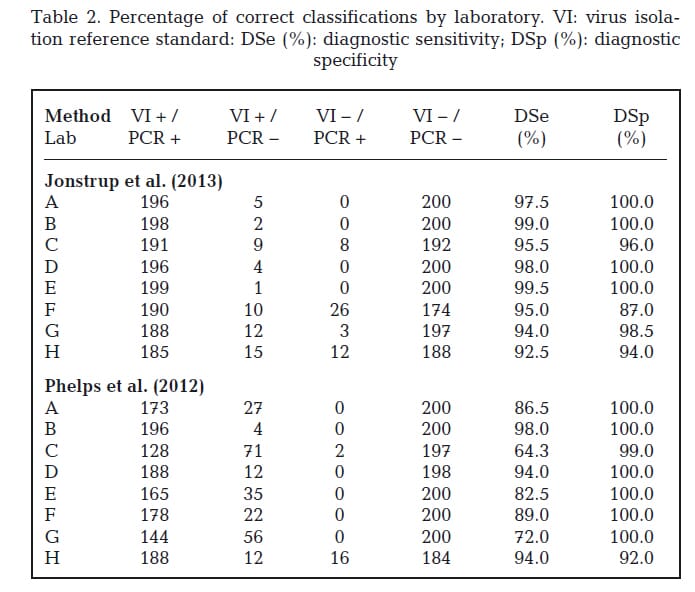
The percentages of correct positive classifications by laboratory for the portion of positive tissue homogenates that tested positive were calculated (Table 2). Out of the 200 samples positive by virus isolation for VHSV, 185 to 199 tested positive by the Jonstrup assay and 128 to 196 tested positive by the Phelps assay.
Sensitivity was estimated to be 0.96 (95% CI: 0.95, 0.97) for the Jonstrup assay and 0.85 (95% CI: 0.83, 0.87) for the Phelps assay. Test results were aligned (Fig. 1) to evaluate whether any particular samples produced false negative test results in multiple laboratories. Three samples (3, 108, and 116) were reported as false negatives when evaluated by the Jonstrup assay by most laboratories.
Other samples with false negative results re ported by 2 or more labora tories included sample 77 (4 laboratories), samples 70 and 81 (3 laboratories), and samples 65, 71, 94, 138 and 160 (2 laboratories). False negative results were reported for both VHSV IVa (5 samples) and VHSV IVb (24 samples).
The number of injected VHSV IVb (2 samples) false negative samples was similar to the number of injected IVa (5 samples) false negative samples. The remaining VHSV IVb false negative samples included 7 natural infection and 15 experimental infection contact animals. An additional 18 samples were reported as false negative by a single laboratory.
The number of virus isolation positive samples with false negative reports when evaluated by the Phelps assay was high (79 samples had at least 1 laboratory reporting them as false negative) as compared to the Jonstrup assay (29 samples). Samples 3, 108, and 116 were also reported as false negatives by the Phelps assay.
Diagnostic specificity
The percentages of negative classifications by laboratory for the portion of negative tissue homogenates that tested negative were calculated (Table 2). Out of the 200 samples negative by virus isolation for VHSV, 174 to 200 tested negative by the Jonstrup assay and 184 to 200 by the
Phelps assay. Specificity was estimated to be 0.97 (95% CI: 0.97, 0.98) for the Jonstrup assay and 0.99 (95% CI: 0.98, 0.99) for the Phelps assay.
Four laboratories (A, B, D, and E) reported no false positive test results for either assay. False positive results included both low and high Ct values.
No single sample was reported as false positive by multiple laboratories. Two labs (F and H) re ported the majority of false positives for the Jonstrup assay; a single lab (H) had the majority of false positives for the Phelps assay. Lab H also experienced a high number of false negatives
with the Jonstrup assay.
Discussion
The objective of this study was to evaluate (side by side) the diagnostic performance of 2 assays under consideration for deployment to a network of independent laboratories that provide surveillance, diagnostic, and fish facility monitoring testing for VHSV.
The tissue samples utilized in this comparison were selected to closely match the types of samples a dia - gnostic laboratory might receive for routine testing in terms of species, sample matrix, attribute being measured, and target concentrations.
In order to reduce but not eliminate inhomogeneity and instability concerns, tissue homogenate supernatants were selected as the testing matrix. Extensive homogeneity and stability testing was not possible due to sample size limitations.
However, in such cases the use of this type of sample is still useful as long as this uncertainty is considered during the evaluation of results (ISO 2010). Estimates of diagnostic sensitivity and specificity from this study are most likely biased as the true classification for each sample is based on an imperfect test.
The trueness of the classification does not preclude comparing assay performance across laboratories on these samples. Further, the reference test is detecting infectious virus, while the PCR assays detect the presence of nucleic acid. The performance (robustness) of the assays was compared across the laboratories using the percentage of samples with the correct classification.
This comparison is also impacted by inherent biases which are identical for all 8 laboratories. Therefore, a comparison of the 2 different rRT-PCR assays performance on the same tissue panel by testing laboratories is not likely to be impacted by these particular inherent biases. In this multi-laboratory evaluation of rRT-PCR assays for VHSV detection, the Jonstrup assay (Jonstrup et al. 2013) outperformed the Phelps assay (Phelps et al. 2012) for diagnostic sensitivity, but assay performances were similar for diagnostic specificity. The Phelps assay is a modification of the Garver et al. (2011) VHSV rRT-PCR 2-step assay to a 1-step approach (Phelps et al. 2012).
Diagnostic sensitivity estimates for the Phelps assay of <90% by 5 out of the 8 participating laboratories was lower than ex - pected based on analytical published data of Phelps et al. (2013) and comparisons of the limit of detection (LOD) on representative isolates from each genotype of VHSV for this assay (Warg et al. 2014).
A comparison of analytical sensitivity for the Phelps assay and the Garver assay reported similar LODs (Warg et al. 2014). However, efficiency estimates for the Phelps assay (102−151% on VHSV MI03 RNA standards) were suboptimal (optimal being 90−104%). Virus isolation is an imperfect standard, and this was taken into consideration when assigning the infection status of a sample and evaluating assay com parison data (Fig. 1).
Virus isolation was re - peated on all tissue homogenate supernatants when thawed for aliquoting into the test panels. Virus was recovered from 134 of the 200 samples that originally tested positive for VHSV. This is not unexpected as material frozen (whole fish, tissue homogenate, or supernatant) and freeze−thaw events can impact the ability to recover replicating virus from a diagnostic sample (Meyers et al. 1999, Arkush et al. 2006, Hervé-Claude et al. 2008), but have less impact on the ability to detect viral RNA (Phelps et al. 2013).
All 200 negative samples tested negative on repeat virus isolation testing. Samples 3, 108, and 116 showed ‘false negative’ results with both PCR assays (Fig. 1) in multiple but not all laboratories. Samples 108 (largemouth bass) and 116 (channel catfish) were both fish cohabited in tanks with injected fish (Table 1) and were negative for virus isolation on repeat testing.
Sample 3 was a naturally infected round goby that was positive for virus isolation on repeat testing. False negative PCR results were obtained by the Jonstrup assay for 6 cohabitant bluegill (samples 65, 70, 71, 77, 81, 84) and 2 injected fish (muskellunge 138 and herring 160); these samples were negative for virus isolation on repeat testing.
Samples reported as false negative by the Phelps assay included wild, cohabitants, and injected fish. False positive classification is equally a concern. The number of false positives was low by both assays. Sample history and a review of samples testing false positive do not suggest misclassification of infection status. On the basis of this study, the rRT-PCR assay of Jonstrup et al. (2013) can be used as a valuable tool when surveillance or suspect VHSV samples are submitted to a laboratory for testing. In addition, the high throughput capacity and the speed of the assay will allow rapid identification of VHSV affected farms or populations.
October 2014




